Data structures
R has multiple data structures. If you are familiar with excel you can think of data structures as building blocks of a table and the table it self, a table is similar to a sheet in excel. Most of the time you will deal with tabular data sets, you will manipulate them, take sub-sections of them. It is good to know what are the common data structures in R and how they can be used.
Vectors
Vectors are one the core R data structures. It is basically a list of elements of the same type (numeric,character or logical). Later you will see that every column of a table will be represented as a vector. R handles vectors easily and intuitively. You can create vectors with c() function, however that is not the only way. The operations on vectors will propagate to all the elements of the vectors.
x <- c(1, 3, 2, 10, 5) #create a vector x with 5 components
x
## [1] 1 3 2 10 5
y <- 1:5 #create a vector of consecutive integers y
y + 2 #scalar addition
## [1] 3 4 5 6 7
2 * y #scalar multiplication
## [1] 2 4 6 8 10
y^2 #raise each component to the second power
## [1] 1 4 9 16 25
2^y #raise 2 to the first through fifth power
## [1] 2 4 8 16 32
y #y itself has not been unchanged
## [1] 1 2 3 4 5
y <- y * 2
y #it is now changed
## [1] 2 4 6 8 10
r1 <- rep(1, 3) # create a vector of 1s, length 3
length(r1) #length of the vector
## [1] 3
class(r1) # class of the vector
## [1] "numeric"
a <- 1 # this is actually a vector length one
Matrices
A matrix refers to a numeric array of rows and columns. You can think of it as a stacked version of vectors where each row or column is a vector. One of the easiest ways to create a matrix is to combine vectors of equal length using cbind(), meaning 'column bind'.
x <- c(1, 2, 3, 4)
y <- c(4, 5, 6, 7)
m1 <- cbind(x, y)
m1
## x y
## [1,] 1 4
## [2,] 2 5
## [3,] 3 6
## [4,] 4 7
t(m1) # transpose of m1
## [,1] [,2] [,3] [,4]
## x 1 2 3 4
## y 4 5 6 7
dim(m1) # 2 by 5 matrix
## [1] 4 2
You can also directly list the elements and specify the matrix:
m2 <- matrix(c(1, 3, 2, 5, -1, 2, 2, 3, 9), nrow = 3)
m2
## [,1] [,2] [,3]
## [1,] 1 5 2
## [2,] 3 -1 3
## [3,] 2 2 9
Matrices and the next data structure data frames are tabular data structures. You can subset them using [] and providing desired rows and columns to subset. Here is how that works conceptually:
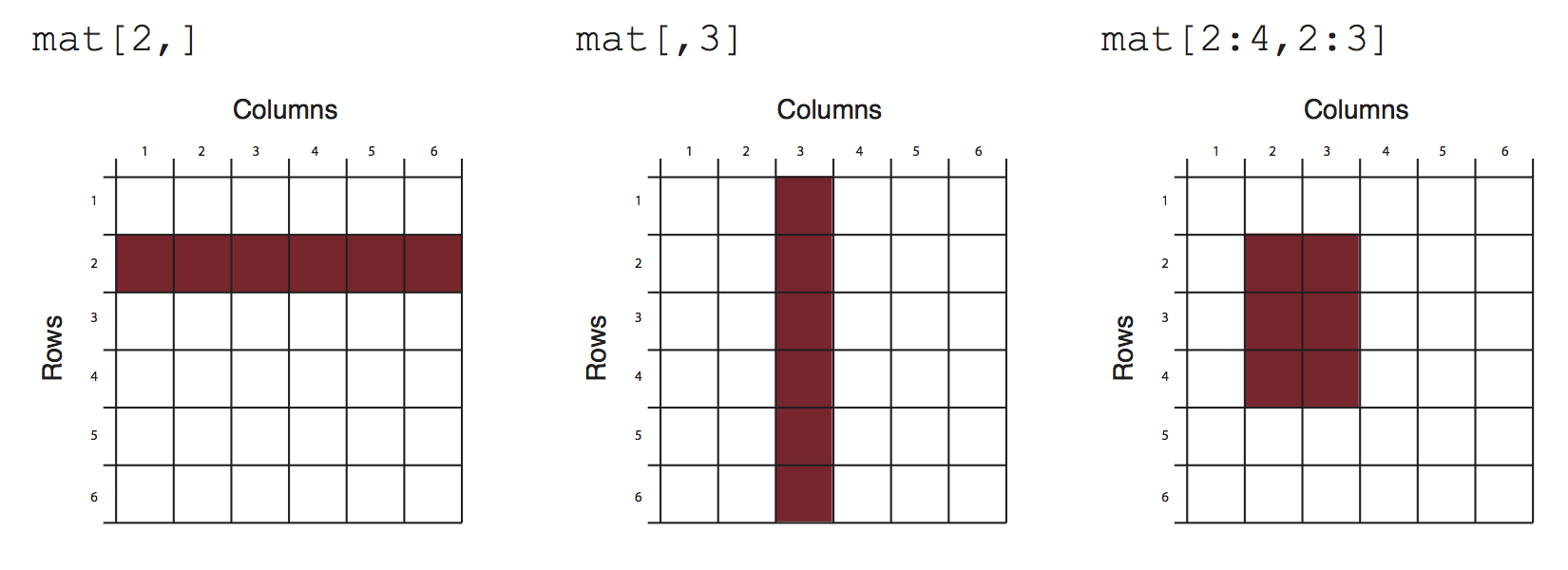 slicing/subsetting of a matrix and data frame.There are additional ways to subset data frames, see the next section.
slicing/subsetting of a matrix and data frame.There are additional ways to subset data frames, see the next section.
Data Frames
A data frame is more general than a matrix, in that different columns can have different modes (numeric, character, factor, etc.). A data frame can be constructed by data.frame() function. For example, we illustrate how to construct a data frame from genomic intervals or coordinates.
chr <- c("chr1", "chr1", "chr2", "chr2")
strand <- c("-", "-", "+", "+")
start <- c(200, 4000, 100, 400)
end <- c(250, 410, 200, 450)
mydata <- data.frame(chr, start, end, strand)
# change column names
names(mydata) <- c("chr", "start", "end", "strand")
mydata # OR this will work too
## chr start end strand
## 1 chr1 200 250 -
## 2 chr1 4000 410 -
## 3 chr2 100 200 +
## 4 chr2 400 450 +
mydata <- data.frame(chr = chr, start = start, end = end, strand = strand)
mydata
## chr start end strand
## 1 chr1 200 250 -
## 2 chr1 4000 410 -
## 3 chr2 100 200 +
## 4 chr2 400 450 +
There are a variety of ways to extract the elements of a data frame. You can extract certain columns using column numbers or names, or you can extract certain rows by using row numbers. You can also extract data using logical arguments, such as extracting all rows that has a value in a column larger than your threshold.
mydata[, 2:4] # columns 2,3,4 of data frame
## start end strand
## 1 200 250 -
## 2 4000 410 -
## 3 100 200 +
## 4 400 450 +
mydata[, c("chr", "start")] # columns chr and start from data frame
## chr start
## 1 chr1 200
## 2 chr1 4000
## 3 chr2 100
## 4 chr2 400
mydata$start # variable start in the data frame
## [1] 200 4000 100 400
mydata[c(1, 3), ] # get 1st and 3rd rows
## chr start end strand
## 1 chr1 200 250 -
## 3 chr2 100 200 +
mydata[mydata$start > 400, ] # get all rows where start>400
## chr start end strand
## 2 chr1 4000 410 -
Lists
An ordered collection of objects (components). A list allows you to gather a variety of (possibly unrelated) objects under one name.
# example of a list with 4 components a string, a numeric vector, a matrix,
# and a scalar
w <- list(name = "Fred", mynumbers = c(1, 2, 3), mymatrix = matrix(1:4, ncol = 2),
age = 5.3)
w
## $name
## [1] "Fred"
##
## $mynumbers
## [1] 1 2 3
##
## $mymatrix
## [,1] [,2]
## [1,] 1 3
## [2,] 2 4
##
## $age
## [1] 5.3
You can extract elements of a list using the [[]] convention using either its position in the list or its name.
w[[3]] # 3rd component of the list
## [,1] [,2]
## [1,] 1 3
## [2,] 2 4
w[["mynumbers"]] # component named mynumbers in list
## [1] 1 2 3
w$age
## [1] 5.3
Factors
Factors are used to store categorical data. They are important for statistical modeling since categorical variables are treated differently in statistical models than continuos variables. This ensures categorical data treated accordingly in statistical models.
features = c("promoter", "exon", "intron")
f.feat = factor(features)
Important thing to note is that when you are reading a data.frame with read.table() or creating a data frame with data.frame() character columns are stored as factors by default, to change this behaviour you need to set stringsAsFactors=FALSE in read.table() and/or data.frame() function arguments.
Data types
There are four comon data types in R, they are numeric, logical, character and integer. All these data types can be used to create vectors natively.
# create a numeric vector x with 5 components
x <- c(1, 3, 2, 10, 5)
x
## [1] 1 3 2 10 5
# create a logical vector x
x <- c(TRUE, FALSE, TRUE)
x
## [1] TRUE FALSE TRUE
# create a character vector
x <- c("sds", "sd", "as")
x
## [1] "sds" "sd" "as"
class(x)
## [1] "character"
# create an integer vector
x <- c(1L, 2L, 3L)
x
## [1] 1 2 3
class(x)
## [1] "integer"